Portfolio
Intrinsically Disordered Proteins (IDPs)
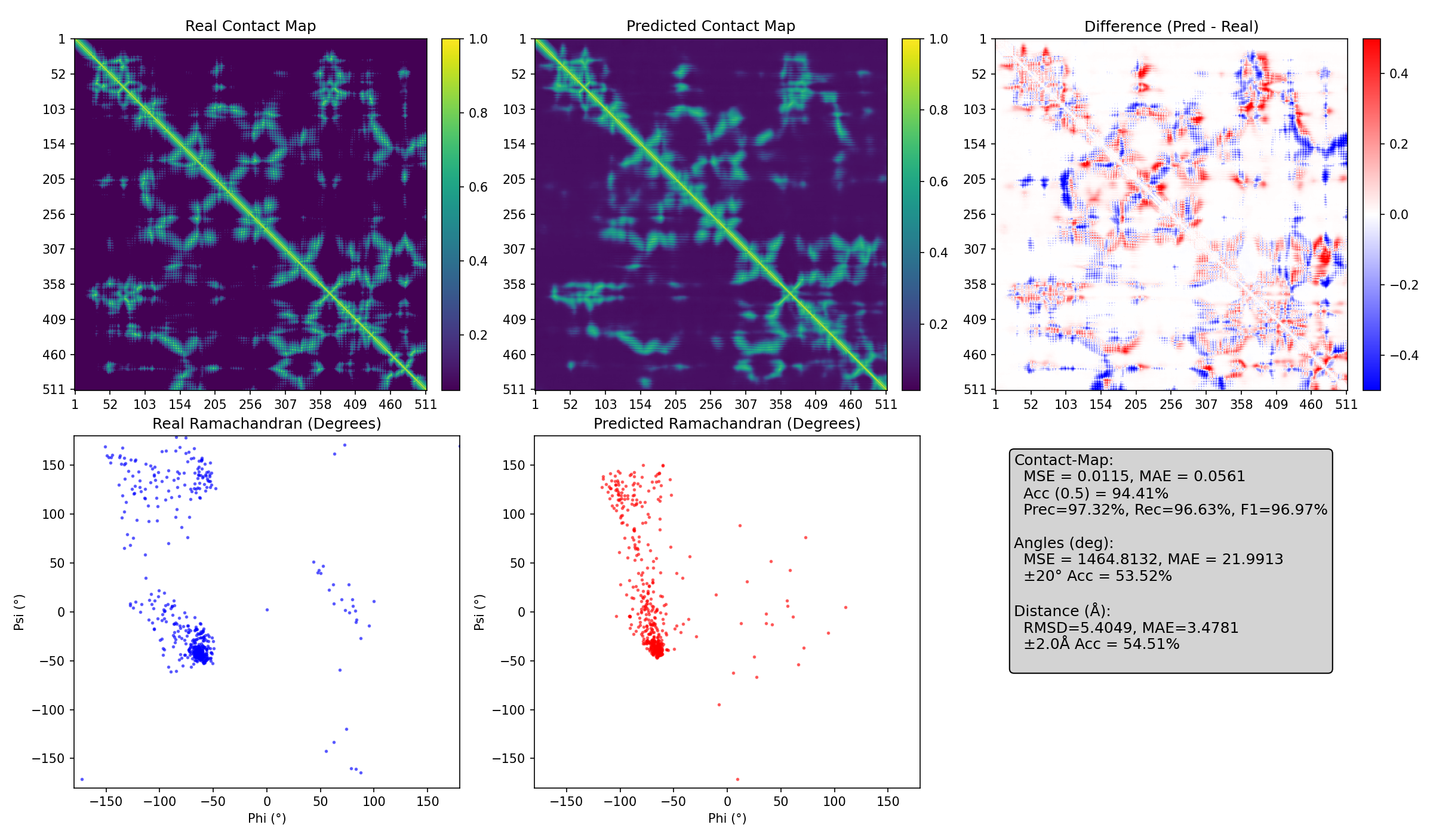
This research explored how phosphorylation regulates the structure and DNA-binding ability of HMGA1, an intrinsically disordered protein (IDP). The work sheds light on the role of IDPs in chromatin remodeling and cancer progression.
Methods
Experimental Methods
- NMR Spectroscopy: Analyzed the conformational ensemble of HMGA1 and characterized its transient structural elements.
- Isothermal Titration Calorimetry (ITC): Measured the DNA-binding affinities of phosphorylated HMGA1.
- Circular Dichroism (CD) spectroscopy to monitor structural changes upon phosphorylation.
Computational Methods
- Molecular dynamics simulations to identify phosphorylation-induced structural changes.
- Bioinformatics analysis of sequence conservation and phosphorylation sites.
- Statistical analysis of NMR chemical shift data to map structural propensities.
Key Findings
- HMGA1 adopts compact, transient structures regulated by phosphorylation.
- CK2 phosphorylation reduces HMGA1's DNA-binding affinity, highlighting its regulatory role in chromatin dynamics.
- Identified key phosphorylation sites as potential therapeutic targets in HMGA1-associated cancers.
- Demonstrated how post-translational modifications can fine-tune IDP function in gene regulation.
Related Publications:
- Kohl, B., Zhong, X., Herrmann, C., Stoll, R. (2019). Phosphorylation orchestrates the structural ensemble of the intrinsically disordered protein HMGA1a and modulates its DNA binding to the NFkB promoter. Nuc. Acids. Res. 47, 11906-11920.
DOI: 10.1093/nar/gkz614
NINJ1 and Membrane Rupture
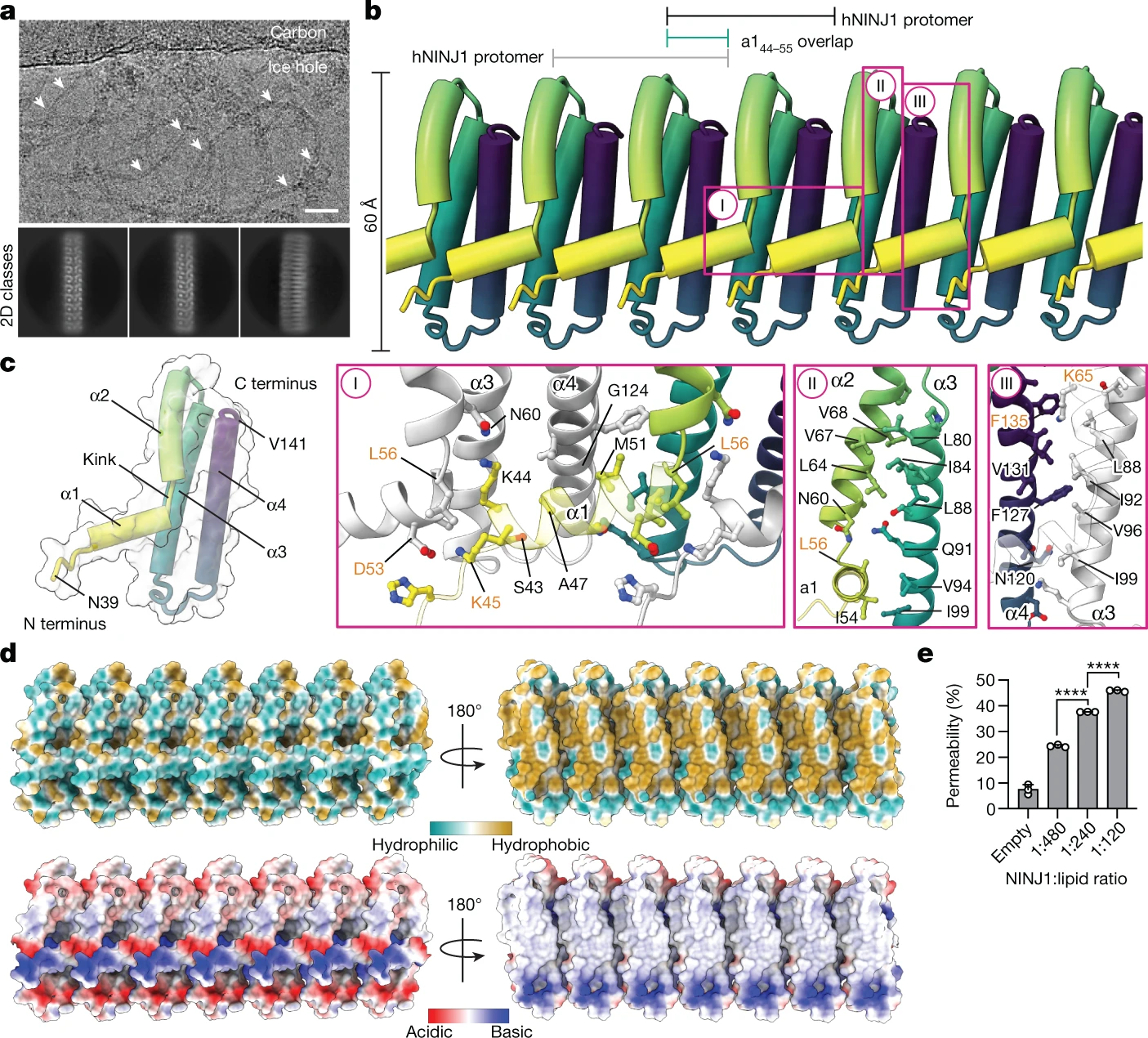
This study revealed the structural basis of NINJ1-mediated plasma membrane rupture during lytic cell death, advancing our understanding of inflammatory pathways and cell death mechanisms.
Methods
Experimental Methods
- Cryo-Electron Microscopy (Cryo-EM): Solved the structure of NINJ1 filaments in various conformational states.
- Super-Resolution Microscopy: Visualized NINJ1 clusters in pyroptotic cells to correlate structural changes with membrane disruption.
- Biochemical assays to characterize NINJ1 oligomerization and membrane interactions.
Computational Methods
- Molecular Simulations: Modeled NINJ1's interaction with membrane edges to understand its rupture mechanism.
- Image processing and 3D reconstruction of cryo-EM data.
- Structural bioinformatics analysis of NINJ1 sequence conservation and evolution.
Key Findings
- NINJ1 forms amphipathic filaments that stabilize and rupture membranes during cell death.
- Identified α-helices critical for filament assembly and membrane binding.
- Demonstrated NINJ1's potential as a therapeutic target in inflammation-related diseases.
- Revealed the molecular basis for NINJ1-mediated membrane disruption in cell death pathways.
Related Publications:
- Degen M., Santos JC., ... Kohl B., et al. (2023). Structural basis of NINJ1-mediated plasma membrane rupture in cell death. Nature 618, 1065–1071.
DOI: 10.1038/s41586-023-05991-z
Photo-Crosslinking and Protein Labeling
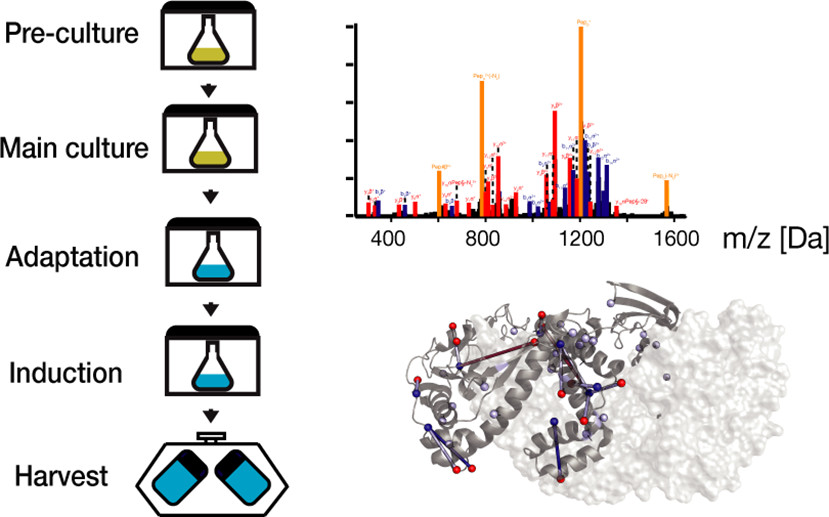
This project developed a high-yield protocol for producing photo-leucine-labeled proteins for cross-linking mass spectrometry (XL-MS), enabling precise mapping of protein-protein interactions in complex biological systems.
Methods
Experimental Methods
- Bacterial Expression: Optimized E. coli strains and growth conditions for photo-leucine incorporation.
- UV Cross-linking: Developed protocols for controlled photo-activation and cross-linking.
- Mass Spectrometry: Implemented targeted approaches for cross-link identification.
Analytical Methods
- HPLC purification and quality control of labeled proteins.
- LC-MS/MS analysis for cross-link identification and quantification.
- Statistical analysis of labeling efficiency and cross-linking yields.
Key Findings
- Achieved unprecedented photo-leucine incorporation rates (>30%) while maintaining high protein yields.
- Developed a cost-effective protocol that reduces isotope labeling expenses by 90%.
- Identified 12 novel cross-linked sites in molecular chaperones, advancing structural mapping.
- Demonstrated broad applicability for studying protein-protein interactions in various systems.
Related Publications:
- Kohl, B., Brüderlin, M., et al. (2020). Protocol for high-yield production of photo-leucine-labeled proteins in Eschericha coli. J. Proteome Res. 19, 8, 3100-3108.
DOI: 10.1021/acs.jproteome.0c00105
Scorpion Toxin: Structure and Bioinformatics
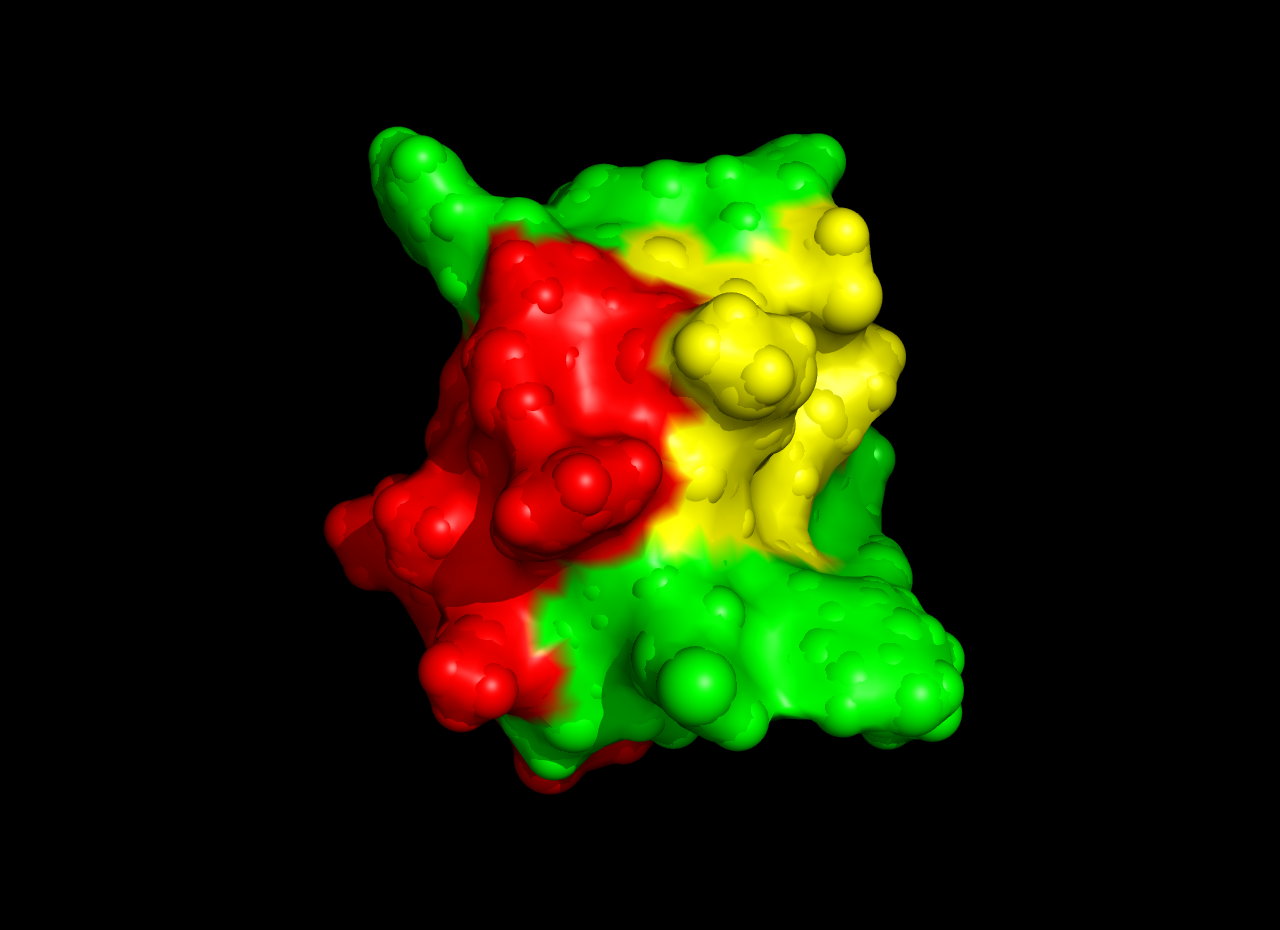
This project focused on the structure and bioinformatics of Bs6, a scorpion-derived toxin with high selectivity for Kv1.3 potassium channels. Combining advanced experimental and computational techniques, this work provides valuable insights into autoimmune disease treatments and the development of channel-specific inhibitors.
Methods
Experimental Methods
- Synthesized Bs6 toxin using solid-phase peptide synthesis with detailed optimization for yield and purity.
- Conducted NMR spectroscopy for the 3D structural determination of Bs6, focusing on its functional motifs and secondary structure.
- Utilized electrophysiological assays on Xenopus oocytes to measure the inhibitory effect of Bs6 on Kv1.3 and Kv7.1 channels.
Computational Methods
- Performed molecular docking to predict Bs6 binding to potassium channels.
- Ran molecular dynamics (MD) simulations using GROMACS to evaluate toxin-channel stability and interactions at the atomic level.
- Conducted sequence alignment and bioinformatics analyses to classify Bs6 into the α-KTx3 family.
Key Findings
- Bs6 adopts a βαββ-fold motif stabilized by three disulfide bonds, providing structural rigidity essential for channel binding.
- Bs6 selectively blocks Kv1.3 potassium channels with an IC50 value of 0.89 nM, showing potential for autoimmune disease treatments.
- Molecular dynamics simulations revealed key residues (e.g., Lys27, Arg9, and Phe25) responsible for stabilizing Bs6's interaction with Kv1.3.
- Bioinformatics analysis classified Bs6 as part of the α-KTx3 family, highlighting its structural and functional similarity to other potent channel inhibitors.
Related Publications:
- Kohl, B., et al. (2015). Solid phase synthesis, NMR structure determination of α-KTx3.8, its in silico docking to Kv1.x potassium channels, and electrophysiological analysis provide insights into toxin-channel selectivity. Toxicon, 101, 70-78.
DOI: 10.1016/j.toxicon.2015.04.018